Support vector machines (SVMs) are powerful yet flexible supervised machine learning algorithm which is used for both classification and regression. But generally, they are used in classification problems. In 1960s, SVMs were first introduced but later they got refined in 1990 also. SVMs have their unique way of implementation as compared to other machine learning algorithms. Now a days, they are extremely popular because of their ability to handle multiple continuous and categorical variables.
Working of SVM
The goal of SVM is to find a hyperplane that separates the data points into different classes. A hyperplane is a line in 2D space, a plane in 3D space, or a higher-dimensional surface in n-dimensional space. The hyperplane is chosen in such a way that it maximizes the margin, which is the distance between the hyperplane and the closest data points of each class. The closest data points are called the support vectors.
The distance between the hyperplane and a data point “x” can be calculated using the formula −
distance = (w . x + b) / ||w||
where “w” is the weight vector, “b” is the bias term, and “||w||” is the Euclidean norm of the weight vector. The weight vector “w” is perpendicular to the hyperplane and determines its orientation, while the bias term “b” determines its position.
The optimal hyperplane is found by solving an optimization problem, which is to maximize the margin subject to the constraint that all data points are correctly classified. In other words, we want to find the hyperplane that maximizes the margin between the two classes while ensuring that no data point is misclassified. This is a convex optimization problem that can be solved using quadratic programming.
If the data points are not linearly separable, we can use a technique called kernel trick to map the data points into a higher-dimensional space where they become separable. The kernel function computes the inner product between the mapped data points without computing the mapping itself. This allows us to work with the data points in the higherdimensional space without incurring the computational cost of mapping them.
Let”s understand it in detail with the help of following diagram −
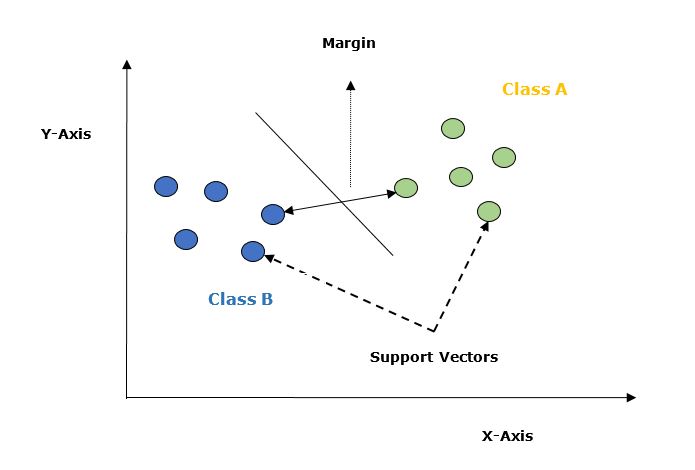
Given below are the important concepts in SVM −
-
Support Vectors − Datapoints that are closest to the hyperplane is called support vectors. Separating line will be defined with the help of these data points.
-
Hyperplane − As we can see in the above diagram it is a decision plane or space which is divided between a set of objects having different classes.
-
Margin − It may be defined as the gap between two lines on the closet data points of different classes. It can be calculated as the perpendicular distance from the line to the support vectors. Large margin is considered as a good margin and small margin is considered as a bad margin.
Implementation in Python
We will use the scikit-learn library to implement SVM in Python. Scikit-learn is a popular machine learning library that provides a wide range of algorithms for classification, regression, clustering, and dimensionality reduction tasks.
We will use the famous Iris dataset, which contains the sepal length, sepal width, petal length, and petal width of three species of iris flowers: Iris setosa, Iris versicolor, and Iris virginica. The goal is to classify the flowers into their respective species based on these four features.
Example
from sklearn.datasets import load_iris
from sklearn.model_selection import train_test_split
from sklearn.svm import SVC
from sklearn.metrics import accuracy_score
# load the iris dataset
iris = load_iris()
# split the data into training and testing sets
X_train, X_test, y_train, y_test = train_test_split(iris.data,
iris.target, test_size=0.2, random_state=42)
# create an SVM classifier with a linear kernel
svm = SVC(kernel=''linear'')
# train the SVM classifier on the training set
svm.fit(X_train, y_train)
# make predictions on the testing set
y_pred = svm.predict(X_test)
# calculate the accuracy of the classifier
accuracy = accuracy_score(y_test, y_pred)
print("Accuracy:", accuracy)
We start by importing the necessary modules from scikit-learn: load_iris to load the iris dataset, train_test_split to split the data into training and testing sets, SVC to create an SVM classifier with a linear kernel, and accuracy_score to calculate the accuracy of the classifier.
We load the iris dataset using load_iris and split the data into training and testing sets using train_test_split. We use a test size of 0.2, which means that 20% of the data will be used for testing and 80% for training. We set the random state to 42 to ensure reproducibility of the results.
We create an SVM classifier with a linear kernel using SVC(kernel=”linear”). We then train the SVM classifier on the training set using svm.fit(X_train, y_train).
Once the classifier is trained, we make predictions on the testing set using svm.predict(X_test). We then calculate the accuracy of the classifier using accuracy_score(y_test, y_pred) and print it to the console.
Output
The output of the code should be something like this −
Accuracy: 1.0
Tuning SVM Parameters
In practice, SVMs often require tuning of their parameters to achieve optimal performance. The most important parameters to tune are the kernel, the regularization parameter C, and the kernel-specific parameters.
The kernel parameter determines the type of kernel to use. The most common kernel types are linear, polynomial, radial basis function (RBF), and sigmoid. The linear kernel is used for linearly separable data, while the other kernels are used for non-linearly separable data.
The regularization parameter C controls the trade-off between maximizing the margin and minimizing the classification error. A higher value of C means that the classifier will try to minimize the classification error at the expense of a smaller margin, while a lower value of C means that the classifier will try to maximize the margin even if it means more misclassifications.
The kernel-specific parameters depend on the type of kernel being used. For example, the polynomial kernel has parameters for the degree of the polynomial and the coefficient of the polynomial, while the RBF kernel has a parameter for the width of the Gaussian function.
We can use cross-validation to tune the parameters of the SVM. Cross-validation involves splitting the data into several subsets and training the classifier on each subset while using the remaining subsets for testing. This allows us to evaluate the performance of the classifier on different subsets of the data and choose the best set of parameters.
Example
from sklearn.model_selection import GridSearchCV
# define the parameter grid
param_grid = {
''C'': [0.1, 1, 10, 100],
''kernel'': [''linear'', ''poly'', ''rbf'', ''sigmoid''],
''degree'': [2, 3, 4],
''coef0'': [0.0, 0.1, 0.5],
''gamma'': [''scale'', ''auto'']
}
# create an SVM classifier
svm = SVC()
# perform grid search to find the best set of parameters
grid_search = GridSearchCV(svm, param_grid, cv=5)
grid_search.fit(X_train, y_train)
# print the best set of parameters and their accuracy
print("Best parameters:", grid_search.best_params_)
print("Best accuracy:", grid_search.best_score_)
We start by importing the GridSearchCV module from scikit-learn, which is a tool for performing grid search on a set of parameters. We define a parameter grid that contains the possible values for each parameter we want to tune.
We create an SVM classifier using SVC() and then pass it to GridSearchCV along with the parameter grid and the number of cross-validation folds (cv=5). We then call grid_search.fit(X_train, y_train) to perform the grid search.
Once the grid search is complete, we print the best set of parameters and their accuracy using grid_search.best_params_ and grid_search.best_score_, respectively.
Output
On executing this program, you will get the following output −
Best parameters: {''C'': 0.1, ''coef0'': 0.5, ''degree'': 3, ''gamma'': ''scale'', ''kernel'': ''poly''}
Best accuracy: 0.975
This means that the best set of parameters found by the grid search are: C=0.1, coef0=0.5, degree=3, gamma=scale, and kernel=poly. The accuracy achieved by this set of parameters on the training set is 97.5%.
You can now use these parameters to create a new SVM classifier and test its performance on the testing set.