Regional maxima refer to a point where the intensity value of pixels is the highest within an image. In an image, regions which form the regional maxima are the brightest amongst all other regions. Regional maxima are also known as global maxima.
Regional maxima consider the entire image, while local maxima only consider a local neighborhood, to find the pixels with highest intensity.
Regional maxima are a subset of local maxima, so all regional maxima are a local maxima but not all local maxima are
regional maxima.
An image can contain multiple regional maxima, but all regional maxima will be of equal intensity. This happens because only the highest intensity value is considered for regional maxima.
Regional Maxima of Image in Mahotas
In Mahotas, we can find the regional maxima in an image using the mahotas.regmax() function. Regional maxima are identified through intensity peaks within an image because they represent high intensity regions.
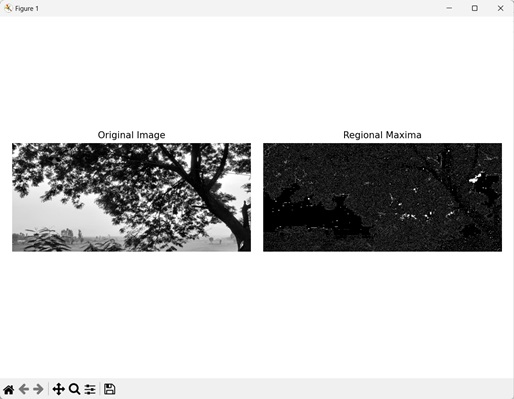
The regional maxima points are highlighted as white while other points are colored in black.
The mahotas.regmax() function
The mahotas.regmax() function extracts regional maxima from an input grayscale image.
It outputs an image where the 1”s represent presence of regional maxima points and 0”s represent normal points.
The regmax() function uses a morphological reconstruction−based approach to find the regional maxima. In this approach, the intensity value of each local maxima region is compared with its neighbors.
If a neighbor is found to have a higher intensity, it becomes the new regional maxima. This process continues until there no region of higher intensity is left, indicating that the regional maxima has been reached.
Syntax
Following is the basic syntax of the regmax() function in mahotas −
mahotas.regmax(f, Bc={3x3 cross}, out={np.empty(f.shape, bool)})
Where,
-
f − It is the input grayscale image.
-
Bc (optional) − It is the structuring element used for connectivity.
-
out(optional) − It is the output array of Boolean data type (defaults to new array of same size as f).
Example
In the following example, we are getting the regional maxima of an image using mh.regmax() function.
import mahotas as mh
import numpy as np
import matplotlib.pyplot as mtplt
# Loading the image
image = mh.imread(''tree.tiff'')
# Converting it to grayscale
image = mh.colors.rgb2gray(image)
# Getting the regional maxima
regional_maxima = mh.regmax(image)
# Creating a figure and axes for subplots
fig, axes = mtplt.subplots(1, 2)
# Displaying the original image
axes[0].imshow(image, cmap=''gray'')
axes[0].set_title(''Original Image'')
axes[0].set_axis_off()
# Displaying the regional maxima
axes[1].imshow(regional_maxima, cmap=''gray'')
axes[1].set_title(''Regional Maxima'')
axes[1].set_axis_off()
# Adjusting spacing between subplots
mtplt.tight_layout()
# Showing the figures
mtplt.show()
Output
Following is the output of the above code −
Using Custom Structuring Element
We can also use a custom structuring element to get the regional maxima from an image. A structuring element is a binary array of odd dimensions consisting of ones and zeroes that defines the connectivity pattern of the neighborhood pixels during image labeling.
The ones indicate the neighboring pixels that are included in the connectivity analysis, while the zeros represent the neighbors that are excluded or ignored.
In mahotas, while extracting regional maxima regions we can use a custom structuring element to define the connectivity of neighboring pixels. We do this by first creating an odd dimension structuring element using the numpy.array() function.
Then, we input this custom structuring element to the Bc parameter in the regmax() function.
For example, let”s consider the custom structuring element: [[0, 0, 0, 0, 1], [0, 0, 1, 0, 0], [1, 0, 0, 0, 0], [0, 0, 0, 1, 0], [0, 1, 0, 0, 0]]. This structuring element implies horizontal connectivity, i.e., only the pixels horizontally left or right of another pixel are considered its neighbors.
Example
In this example, we are using a custom structuring element to get the regional maxima of an image.
import mahotas as mh
import numpy as np
import matplotlib.pyplot as mtplt
# Loading the image
image = mh.imread(''sun.png'')
# Converting it to grayscale
image = mh.colors.rgb2gray(image)
# Setting custom structuring element
struct_element = np.array([[0, 0, 0, 0, 1],
[0, 0, 1, 0, 0],
[1, 0, 0, 0, 0],
[0, 0, 0, 1, 0],
[0, 1, 0, 0, 0]])
# Getting the regional maxima
regional_maxima = mh.regmax(image, Bc=struct_element)
# Creating a figure and axes for subplots
fig, axes = mtplt.subplots(1, 2)
# Displaying the original image
axes[0].imshow(image, cmap=''gray'')
axes[0].set_title(''Original Image'')
axes[0].set_axis_off()
# Displaying the regional maxima
axes[1].imshow(regional_maxima, cmap=''gray'')
axes[1].set_title(''Regional Maxima'')
axes[1].set_axis_off()
# Adjusting spacing between subplots
mtplt.tight_layout()
# Showing the figures
mtplt.show()
Output
Output of the above code is as follows −

Using a Specific Region of an Image
We can also find the regional maxima of a specific region of an image. A specific region of an image refers to a small part of a larger image. The specific region can be extracted by cropping the original image to remove unnecessary areas.
In mahotas, we can find the regional maxima within a portion of an image. First, we crop the original image by specifying the required dimensions of the x and y axis. Then we use the cropped image and get the regional maxima using the regmax() function.
For example, let’s say we specify [:800, 70:] as the dimensions of x and y axis respectively. Then, the cropped image will be in range of 0 to 800 pixels for x−axis and 70 to max dimension for y−axis.
Example
In this example, we are getting the regional maxima within a specific region of an image.
import mahotas as mh
import numpy as np
import matplotlib.pyplot as mtplt
# Loading the image
image = mh.imread(''nature.jpeg'')
# Converting it to grayscale
image = mh.colors.rgb2gray(image)
# Using specific region of the image
image = image[:800, 70:]
# Getting the regional maxima
regional_maxima = mh.regmax(image)
# Creating a figure and axes for subplots
fig, axes = mtplt.subplots(1, 2)
# Displaying the original image
axes[0].imshow(image, cmap=''gray'')
axes[0].set_title(''Original Image'')
axes[0].set_axis_off()
# Displaying the regional maxima
axes[1].imshow(regional_maxima, cmap=''gray'')
axes[1].set_title(''Regional Maxima'')
axes[1].set_axis_off()
# Adjusting spacing between subplots
mtplt.tight_layout()
# Showing the figures
mtplt.show()
Output
After executing the above code, we get the following output −
